Research Areas
The broad research interests of our laboratory focus on understanding the molecular pathology of cancer development and progression, in hopes that this will lead to better treatment for patients. We use molecular biology, protein biochemistry, cell biology, and animal models to address relevant questions. Much of our work is conducted in the context of the hematopoietic system, through studies in cancer cell lines and in mice.
Currently, our work is in two distinct areas:
1) The function of transcription factor AML1, and its leukemogenic fusion protein AML1-ETO, in the contexts of normal blood cell development and leukemia.
2) Protein modification by a ubiquitin-like molecule, ISG15, and the functions of ISG15-specific protease, UBP43, in interferon signaling and in cancer.
3) RNA processing such as primary RNA splicing to mRNA and polyadenylation in hematological disease.
Transcription Factor RUNX1 in Blood Cells and Related Human Diseases
RUNX1, also known as AML1, CBFα2, and PEBP2α2, is a member of the Runt family of proteins. The major function of RUNX1 is to operate as a DNA-binding transcription factor. Studies over the past 20 years have established RUNX1 as a critical player in specification of hematopoietic stem cells and further blood cell differentiation. The importance of RUNX1 is further validated by its prevalence in a variety of hematologic diseases and malignancies, including myelodysplastic syndrome, myeloproliferative neoplasms, and multiple forms for leukemia. As a transcription factor, RUNX1 binds to the specific DNA sequence in regulatory regions and interact with other transcription regulators to guide the expression of its target genes. We are interested in the identification of RUNX1 regulated genes and characterization of their role in hematopoiesis and related disease development.

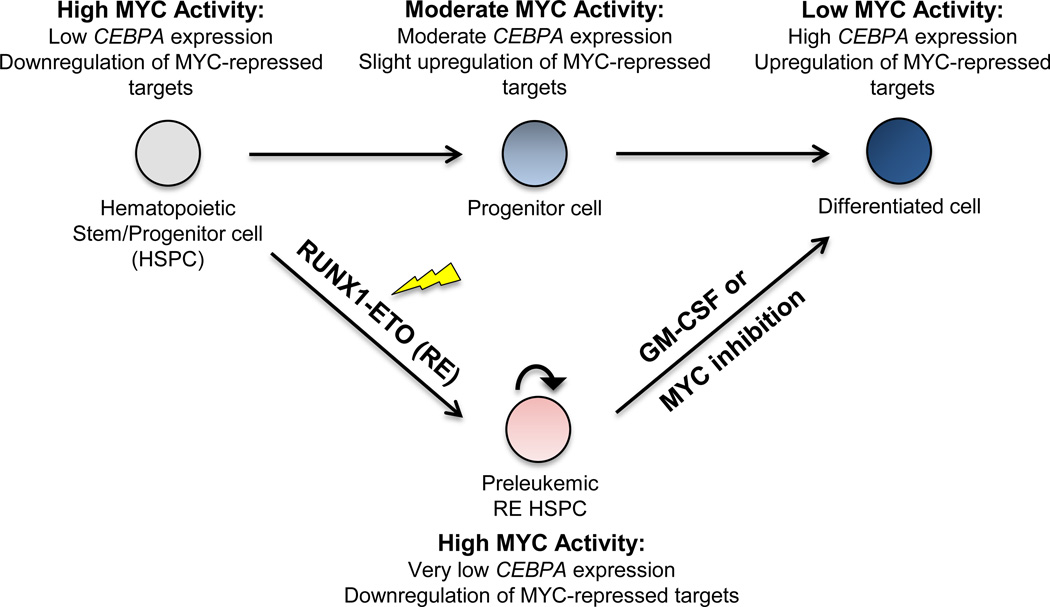
RUNX1-ETO and Acute Myeloid Leukemia
Acute myeloid leukemia (AML) is the most prevalent form of adult leukemia. Chromosomal translocations are found in over 30% of AML, the most common of which is a translocation between chromosome 8 and chromosome 21, t(8;21)(q22;q22), occurring in up to 40% of AML cases categorized within the French-American-British (FAB) subtype M2. This translocation results in the expression of fusion protein RUNX1-ETO. Although sufficient for in vitro immortalization, RUNX1-ETO requires additional cooperating mutations to induce leukemia in vivo. RUNX1-ETO functions as a transcriptional repressor through its recruitment of the corepressors N-CoR and SMRT and their associated histone deacetylases. RUNX1-ETO can also activate gene expression via its association with transcription activators. We are interested in molecular pathology of the RUNX1-ETO mediated leukemogenesis using mouse models and in therapeutic options based on alterations of related gene expression.
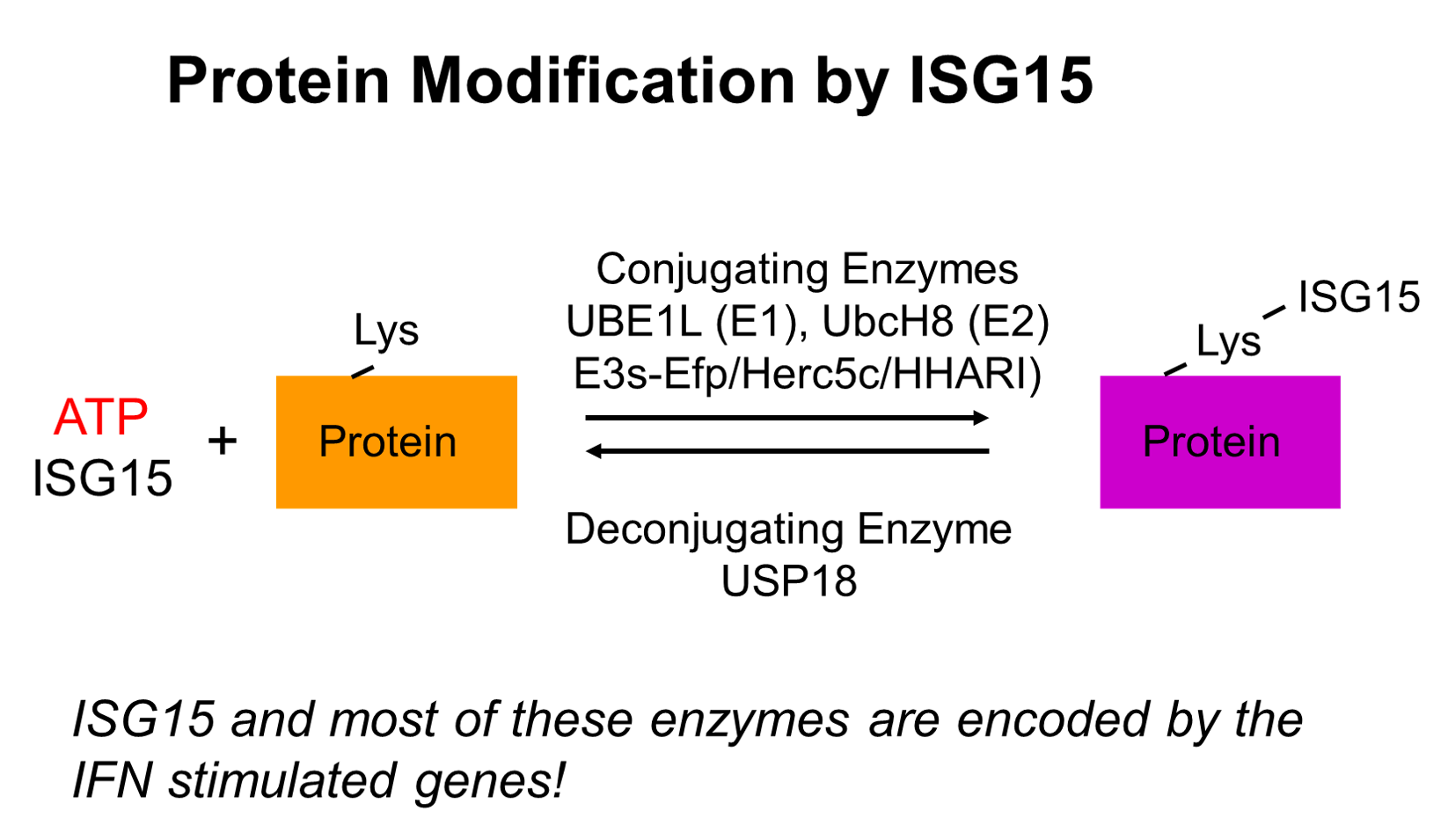
ISG15 and Protein ISGylation in Cancer Immunity
Interferons are a group of cytokines that play important roles during immune responses. Interferons activate interferon stimulated gene (ISG) expression via specific cell surface receptors and JAK/STAT signal transduction pathways. ISG15 gene is highly upregulated upon interferon stimulation. ISG15 protein is a ubiquitin like modifier and forms conjugates with other proteins (ISGylation). A series of distinct enzymes are involved in the process of protein ISGylation, including ubiquitin-activating enzyme E1 homolog (UBE1L), a conjugating enzyme, (UBCH8) (E2), protein ligases (E3), and an ISG15-specific protease USP18, as well as some viral proteins. Interestingly, ISG15 and the majority of its modification enzymes are encoded by ISGs, suggesting that protein ISGylation is a tightly regulated process and plays important roles in immune responses. Protein ISGylation is strongly enhanced upon pathogen infection and cellular stress related to interferon induction. We are interested in how protein ISGylation regulation affects human cancer development.
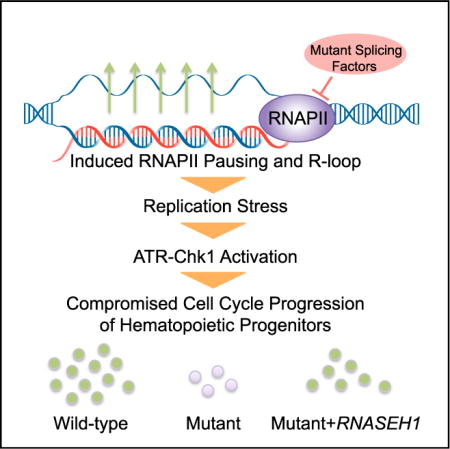
RNA Processing in Hematological Disease
RNA processing, such as primary RNA splicing to mRNA, 5’ cap addition, 3’ polyadenylation, and methylation, play critical roles in modulating gene expression. Mutations in several general splicing factors have been discovered in myelodysplastic syndromes (MDS), leukemia, and solid tumors. In addition to dysregulation of splicing, we report a chain of events triggered by multiple splicing factor mutations, especially high-risk alleles in SRSF2 and U2AF1, including elevated R-loops, replication stress, and activation of the ataxia telangiectasia and Rad3-related protein (ATR)-Chk1 pathway. We are interested in studying cooperative mutations of splicing factors with RUNX1 in hematological diseases and analyzing molecular events related to RNA splicing and polyadenylation in leukemia.
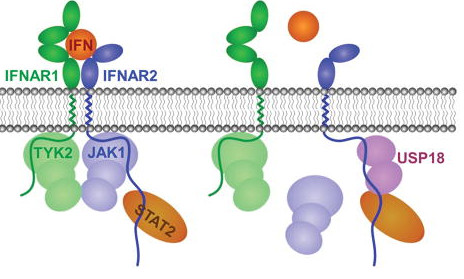
USP18 in Protein Modification and Interferon Signaling
During the analysis of genes differentially expressed between wild type and RUNX1-ETO knock-in mice, we cloned a novel gene product and named it Ubp43 (current nomenclature – Usp18, ubiquitin specific protease 18). USP18 expression is highly induced by interferons and many stress stimulations such as pathogen infection and cancer. Further studies in our lab demonstrate that USP18 is a bona fide ISG15 specific protease. USP18 deficient cells have higher levels of ISGylated proteins than wild type cells. More interestingly, in addition to its function is regulation of protein modification by ISG15, USP18 is a potent inhibitor of Type I and Type III interferon signaling independent of its enzyme activity. We are interested in understanding the molecular basis of its functions in immune responses and exploring related therapeutic potential.